Help
Main menu
Click on the "hamburguer" icon to expand the menu. As shown in the image below, there are links to pages which may be of your interest, including this "Help" page and an explanation of how the Spongilla phylome was constructed.

Searching the phylome
Use the input box to type words searching for trees. Those words will be searched among the tree ID and name, and tree leaves data including proteins IDs and names, automated names, curated names, eggnog-mapper OGs, and PFAM domains. Note that an "AND" logical operation is performed, so that if you search for several words, only those trees with hits from all the words will be reported.
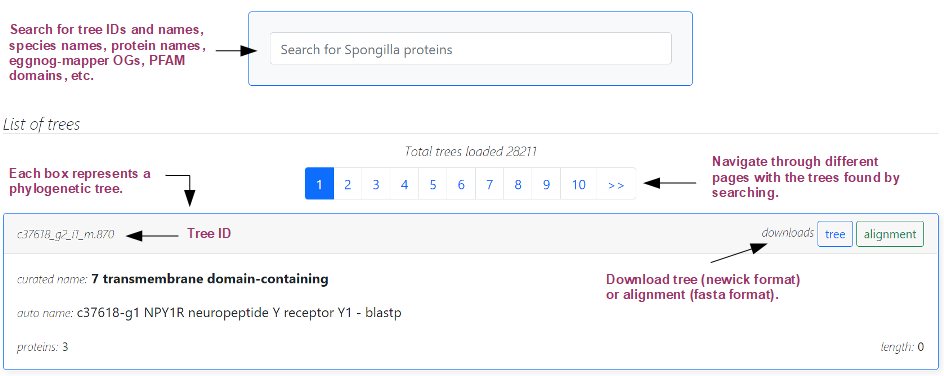
As shown in the image above, a list of trees found will be reported. The total number of trees found is shown, along with a page navigator, since not every tree is shown at once. Each box represents one found tree, and includes a header with the tree ID and buttons to download the newick tree and the fasta alignment, and a body with tree information, including curated (when available) and automated name of the seed protein of the tree, along with the number of proteins and the alignment length. Click on a tree box to visualize it.
Understanding trees
Once you click on a tree box from the search results page, only the box with data from the selected tree will be shown, with an additional "back" button to go back to the search results page.
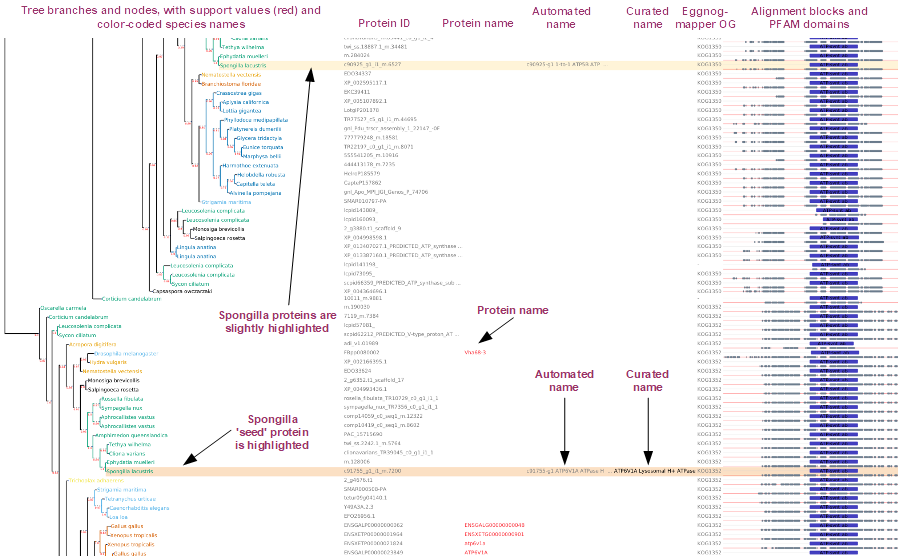
As shown in the image below, the default layout of a tree shows different data.
Towards the left we show the tree branches and nodes. Branches show a support value (in red), and nodes show the species name of the protein, with a code of colors per species and taxonomic group.
Next, we show tabulated data, with different names and identifiers, which are empty for those proteins lacking such info. First we show the proteins ID in gray; second, in red, the protein name, when available; next, the automated name (gray) and curated name (black, when available); and finally the eggnog-mapper Orthologous Group.
Finally, to the right end of the image, we show the multiple sequence alignment, including the aligned (gray) and unalinged (red) blocks, and the PFAM domains identified for each protein.
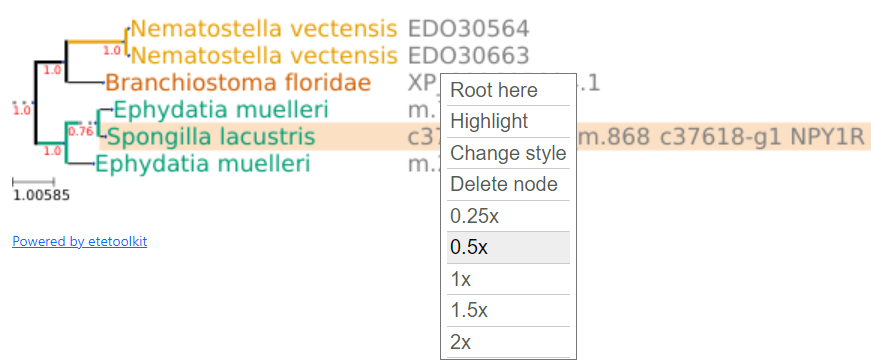
Additional actions on trees
By clicking on a tree node a menu will popup with several additional options. "Root here" allows rooting the tree on the node that was clicked to open the popup menu. "Highlight" allows highlighting/unhighlighting the clicked node. "Change style" allows showing the default ete3 layout for a tree, specially to show the multiple sequence alignment for every residue. "Delete node" allows deleting a node. Finally, there are 5 zoom options, which actually are means to render the tree in smaller or larger image resolutions.